Research
MycoTransport
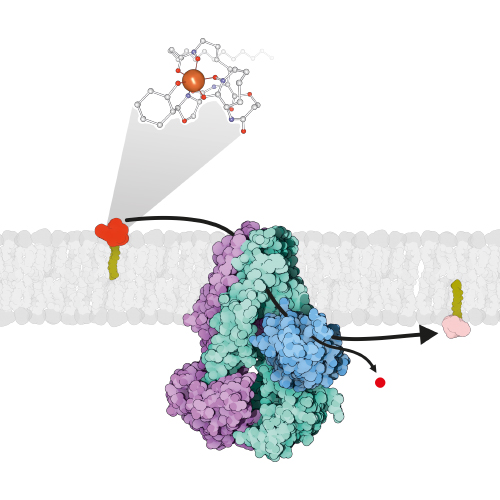
Mycobacterium tuberculosis is one of the best studied bacteria (nearly 80’000 entries on pubmed). However, due to its complex cell wall, membrane transport processes are poorly understood in this important pathogen, which is responsible for the death of more than 2 million people each year. We investigate mycobacterial transporters involved in the shuttling of siderophores, small organic compounds that are produced by the bacillus to steal iron from the infected host. A recent highlight was the structural and functional characterization of the unusual heterodimeric ABC exporter IrtAB, which imports iron-loaded siderophores across the inner membrane and reduces it in the cytoplasm by virtue of a fused siderophore interacting domain. The biosynthesis of the unique mycobacterial cell wall is a further research area that caught our interest. We work with a proton-driven transporter called Rv1410, which plays an important role in the transport of triacylglycerol and highly glycosylated lipids unique to mycobacteria, called lipoarabinomannans.
DrugEfflux
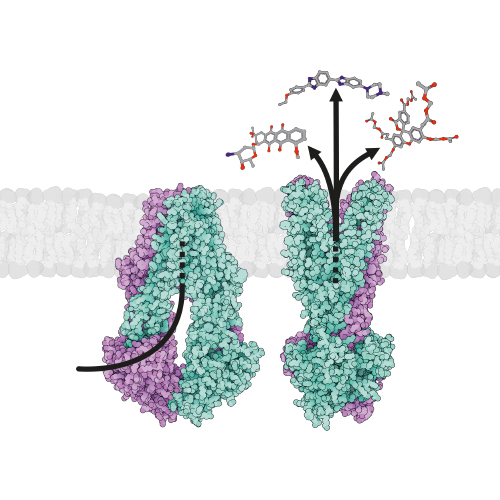
Drug efflux is a common resistance mechanism found in bacteria and cancer cells. Our lab has investigated the structure and function of ABC exporters involved in the efflux of drugs for more than a decade. ABC exporters minimally consist of two transmembrane domains (TMDs) responsible for substrate translocation, which are coupled to a pair of nucleotide binding domains (NBDs) acting as ATP-fuelled motor domains to drive the transport cycle. Current work focusses on a heterodimeric ABC exporter called EfrCD, which strongly contributes to drug efflux in the opportunistic pathogen Enterococcus faecalis. To identify molecular hallmarks of drug efflux pumps, we perform deep mutational scanning to obtain a global picture of residues playing a role in drug recognition and transport. Our long-term goal is to understand promiscuity of drug recognition and the mechanics of drug pumping at the molecular level.
Sybodies
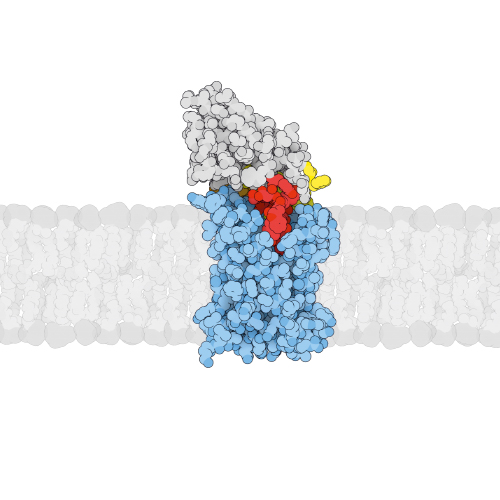
Membrane transporters are intrinsically flexible, which often impedes structural and biochemical studies. This is why there is a great need for affinity reagents that can trap membrane transporters in defined conformational states. We have established a platform for the in vitro selection of synthetic nanobodies called sybodies against challenging membrane proteins. At the heart of the platform are three sybody libraries exhibiting different randomized shapes. A steady stream of publications demonstrates how sybodies increase the thermal stability of labile transporters, trap defined conformations and facilitate X-ray structure determination. We have sent out the sybody library to more than twenty academic research labs around the globe, and we trained a total of eight researchers from Switzerland/Europe/Asia on sybody selections. As a fast response to the Corona crisis, we generated sybodies against the SARS-CoV-2 receptor-binding domain and published the sequences in the open domain.
NestLink
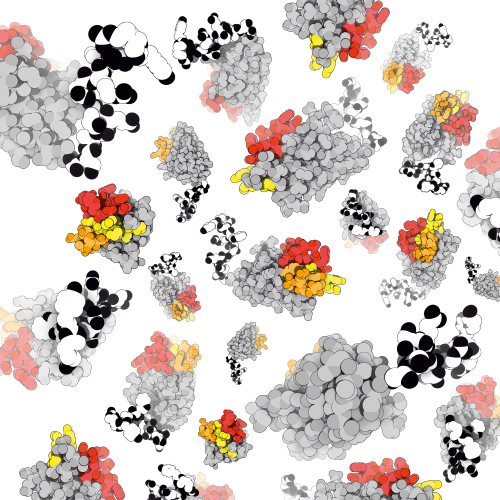
We developed NestLink, an innovative technological concept that combines the benefits of screens and selections. The technology centres around the so called flycodes – genetically encoded peptide barcodes designed for optimal detection by mass spectrometry (LC-MS). Through a combination of state-of-the-art LC-MS and next generation sequencing, we process thousands of library members as an ensemble, while still allowing for characterization of individual candidate molecules within the processed pool of proteins. Hence, we invented a novel protein selection principle that operates in the absence of a classical genotype-phenotype linkage. This paradigm-shift enables novel protein engineering applications, which operate independently of large display particles such as phages and ribosomes. We currently use the flycode technology to deep-mine large pools of nanobodies and sybodies for binders that recognize outer membrane proteins in the context of the bacterial cell with the aim to develop rapid diagnostics and identify novel antibiotics.
